Abstract
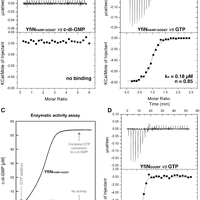
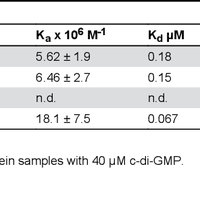
Pseudomonas aeruginosa is responsible for a plethora of biofilm mediated chronic infections among which cystic fibrosis pneumonia is the most frightening. The long-term survival strategy of P. aeruginosa in the patients lungs is based on a fine balance of virulence vs dormant states and on genetic adaptation, in order to select persistent phenotypes as the small colony variants (SCVs), which strongly correlate with antibiotic resistance and poor lung function. Recent studies have coupled SCV with increased levels of the signaling molecule cyclic di-GMP, and demonstrated the central role of the diguanylate cyclase YfiN, part of the tripartite signaling module YifBNR, in c-di-GMP dependent SCV regulation. YfiN, also called TpbB, is a multi-domain membrane enzyme connecting periplasmic stimuli to cytosolic c-di-GMP production by an allosteric inside-out signaling mechanism that, due to the lack of structural data, is still largely hypothetical. We have solved the crystal structure of the catalytic domain (GGDEF), and measured the enzymatic activity of the cytosolic portion in real-time by means of a newly developed method. Based on these results we demonstrate that, unlike other diguanylate cyclase, YfiN does not undergo product feedback inhibition, and that the presence of the HAMP domain is required for dimerization and catalysis. Coupling our structural and kinetic data with an in silico study we are now able to propose a model for the allosteric regulation of YfiN. © 2013 Giardina et al.
Figures
Register to see more suggestions
Mendeley helps you to discover research relevant for your work.
Cite
CITATION STYLE
Giardina, G., Paiardini, A., Fernicola, S., Franceschini, S., Rinaldo, S., Stelitano, V., & Cutruzzolà, F. (2013). Investigating the allosteric regulation of YfiN from Pseudomonas aeruginosa: Clues from the structure of the catalytic domain. PLoS ONE, 8(11). https://doi.org/10.1371/journal.pone.0081324

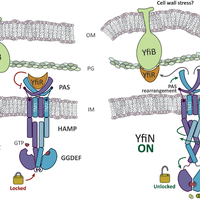
![Figure 1. YfiBNR tripartite system organization. Schematic representation of the localization the YfiBNR system. YfiN is repressed by the specific interaction of YfiR with its periplasmatic domain, while dissociation of the complex, and the consequent activation of YfiN, may be induced by a YfiB-mediated cell wall stress sensing mechanism and/or by redox driven misfolding of YfiR [20].](https://s3-eu-west-1.amazonaws.com/com.mendeley.prod.article-extracted-content/images/2e0c3760-2e95-3a48-ac89-7bff0aef360d/thumbnail-976f810f-a42c-48b0-9c96-4d00e5e51930-0.png)
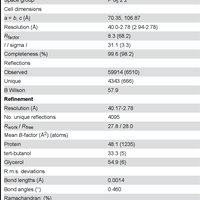
![Figure 2. Cristal structure of YfiNGGDEF. A) Cartoon representation of the YfiNGGDEF structure. The active site and primary inhibitory site (Ip) signature residues (GGDEF and RxxD) are shown in green and magenta respectively. B) Sequence alignment of the GGDEF domain of YfiN with the other DGCs of known structure; PleD from C. crescentus [27,28]; WspR from P. aeruginosa [29]; A1U3W3 from M. aquaeolei [32] and XCC4471 from X. campestris [31]. C) Structure superposition of YfiNGGDEF with the other DGC. YfiNGGDEF (black); PleD from C. crescentus [27,28] (grey - PDB: 2wb4 – rmsd: 1.23 Å); WspR from P. aeruginosa [29] (cyan - PDB: 3i5a - rmsd: 1.31 Å); XCC4471 from X. campestris [31] (light purple - PDB: 3qyy - rmsd: 1.64 Å) and A1U3W3 from M. aquaeolei [32] (dark purple - PDB: 3ign - rmsd: 1.34 Å).](https://s3-eu-west-1.amazonaws.com/com.mendeley.prod.article-extracted-content/images/2e0c3760-2e95-3a48-ac89-7bff0aef360d/thumbnail-30dd4851-c85d-4d52-9054-27ea3538f942-2.png)
![Figure 3. YfiN displays a degenerated Is-Site. A) Binding mode of dimeric c-di-GMP to the I-site of DGCs or to receptor proteins. The first row shows the homo-domain cross-linking (GGDEF/GGDEF), while the second shows the hetero-domain cross-linking (within the same chain) of inhibited PleD and two c-di-GMP receptors. For all structures different colors are used to illustrate domains belonging to different subunits, the side chains of the two arginines and the aspartic acid (R1; R2 and D) are shown as sticks, while the two bound c-di-GMP molecules as balls and sticks. Grey continuous lines indicate H-bonds, while green continuous lines highlight the π-cation interaction among a charged nitrogen atom of the arginine residues and the guanine delocalised πsystem. Ip and Is indicate primary and secondary inhibitory sites respectively. Starting from top left, the reported structure are: PleD (PDB: 2v0n [28]); WspR (PDB: 3bre [30]); A1U3W3 (PDB: 3ign [32]); PleD (PDB: 1w25 [27]); PelD (PDB: 4dn0 ) [33]. and PP4397 (PDB: 3kyf [34]). B) Comparison of the I-site of YfiN and (PDB: 2v0n [28]). The two subunits of a hypothetical inhibited dimer of YfiN (superposed on the structure of PleD) are shown in white and pink, while the same color code of panel A is used for PleD. C-diGMP molecules (bound to PleD) are shown as lines. YfiN lacks two of the three arginine residues binding to c-di-GMP through the stair motif interaction (D273 and N351 - bold labels). Moreover, the presence of a bulky side chain (Y379) yields a shift of helix-A, implying a reduced, sub optimal, volume of the I-site.](https://s3-eu-west-1.amazonaws.com/com.mendeley.prod.article-extracted-content/images/2e0c3760-2e95-3a48-ac89-7bff0aef360d/thumbnail-a212fa50-c78a-4a35-b42f-435e305e16b7-3.png)
![Figure 5. Dimeric model of YfiN. Predicted domain organization of YfiN along with the most significant structural templates found, according to two different fold prediction servers (i.e., Phyre2 [25] and HHPRED [26]) used for homology modeling. The final model including the crystal structure of the catalytic domain is also shown.](https://s3-eu-west-1.amazonaws.com/com.mendeley.prod.article-extracted-content/images/2e0c3760-2e95-3a48-ac89-7bff0aef360d/thumbnail-a9db2dbf-b286-42c9-bddb-aee80e5c582e-6.png)