Abstract
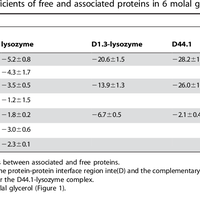
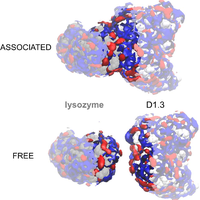
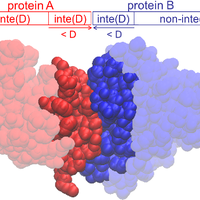
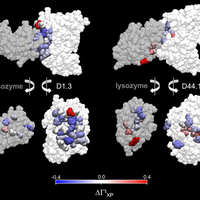
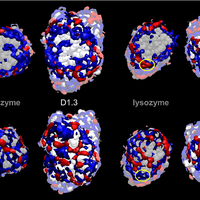
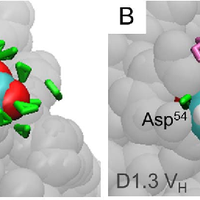
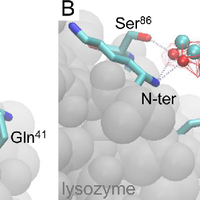
Although the nature of solvent-protein interactions is generally weak and non-specific, addition of cosolvents such as denaturants and osmolytes strengthens protein-protein interactions for some proteins, whereas it weakens protein-protein interactions for others. This is exemplified by the puzzling observation that addition of glycerol oppositely affects the association constants of two antibodies, D1.3 and D44.1, with lysozyme. To resolve this conundrum, we develop a methodology based on the thermodynamic principles of preferential interaction theory and the quantitative characterization of local protein solvation from molecular dynamics simulations. We find that changes of preferential solvent interactions at the protein-protein interface quantitatively account for the opposite effects of glycerol on the antibody-antigen association constants. Detailed characterization of local protein solvation in the free and associated protein states reveals how opposite solvent effects on protein-protein interactions depend on the extent of dewetting of the protein-protein contact region and on structural changes that alter cooperative solvent-protein interactions at the periphery of the protein-protein interface. These results demonstrate the direct relationship between macroscopic solvent effects on protein-protein interactions and atom-scale solvent-protein interactions, and establish a general methodology for predicting and understanding solvent effects on protein-protein interactions in diverse biological environments. © 2013 Vagenende et al.
Figures
Register to see more suggestions
Mendeley helps you to discover research relevant for your work.
Cite
CITATION STYLE
Vagenende, V., Han, A. X., Pek, H. B., & Loo, B. L. W. (2013). Quantifying the Molecular Origins of Opposite Solvent Effects on Protein-Protein Interactions. PLoS Computational Biology, 9(5). https://doi.org/10.1371/journal.pcbi.1003072

![Figure 1. Opposite effects of glycerol on the association constant KA of Fab D44.1 and scFv D1.3 with lysozyme. KA/ KA,0 is the ratio of the association constants with and without glycerol. The data point marked with an asterisk is derived from Goldbaum et al. [21] and all other data points are determined by surface plasmon resonance. doi:10.1371/journal.pcbi.1003072.g001](https://s3-eu-west-1.amazonaws.com/com.mendeley.prod.article-extracted-content/images/3e5a791d-3626-3235-a2b3-f095fac0a495/thumbnail-6380ac91-bbba-464e-93aa-ee127d4b26f2-0.png)